About
Welcome to the lab page for Stephen A. Smith's lab at the University of Michigan, Ann Arbor. You can find out more information about the people in the lab here and the projects here. A lot of the research in the lab focuses on plant evolution, detecting and describing large scale patterns of evolution, examining differences in the rate of molecular evolution, and using new data sources like transcriptomes and genomes to address these questions. A great deal of this involves the development of new methods and new computational tools. My faculty page is here.
Lab updates
-
More updates coming
I have been really bad at keeping these pages up to date over the last couple of yearse (COVID and all). But we are updating all the pages here over the next few days! It has already started with the people page but publications and projects will also be updating shortly.
-
Ericales position
I am pleased to announce that the lab is advertising for a postdoctoral position related to the phylogenomics and evolution of the Ericales plant clade. The position has funding for two years. We are looking to have someone start by the beginning of the year 2020 if possible though there is some flexibility.
Our project, working with the University of Florida, is to collect, sequence, and analyze hundreds of transcriptomes of Ericales along with thousands of targeted capture sequences (we will assist UoF with this). Once these are collected we are interested in answering questions about the evolution of the group between temperate and tropical areas and how morphological evolution intersects with these movements. Position duties include managing the sequencing of transcriptomes and the analysis once the data come back. There will be opportunities to train undergrads and grads as well as direct involvement in project planning. There are also opportunities to help develop, plan, and teach a bilingual workshop on phylogenetics and genomics.
Qualifications include a PhD in biological sciences, preferably with a focus on ecology and on organismal and evolutionary biology. In addition, expertise in laboratory work, organization and analysis of diverse datatypes, and interests in large scale questions regarding the evolution of flowering plants.
The Smith lab is a diverse bunch of folks that range in interests from plant evolution to computational biology. There are great opportunities for interaction and collaboration for those interested. The University of Michigan also has great resources for professional development.
If you are interested or have any questions, please send your CV and information for references (eebsmith@umich.edu).
Review of applications will begin immediately.
The University of Michigan is an equal opportunity employer.
-
PyPHLAWD published
-
Joe Walker defends
Dr. Joe Walker successfully defended his dissertation today! Joe Walker is the first Smith lab graduate student to finish. And he did it in a cool 4 years with all chapters published or in review. Congrats Joe!!

-
PyPHLAWD site up
-
A couple new papers: plant tree of life and quartet sampling
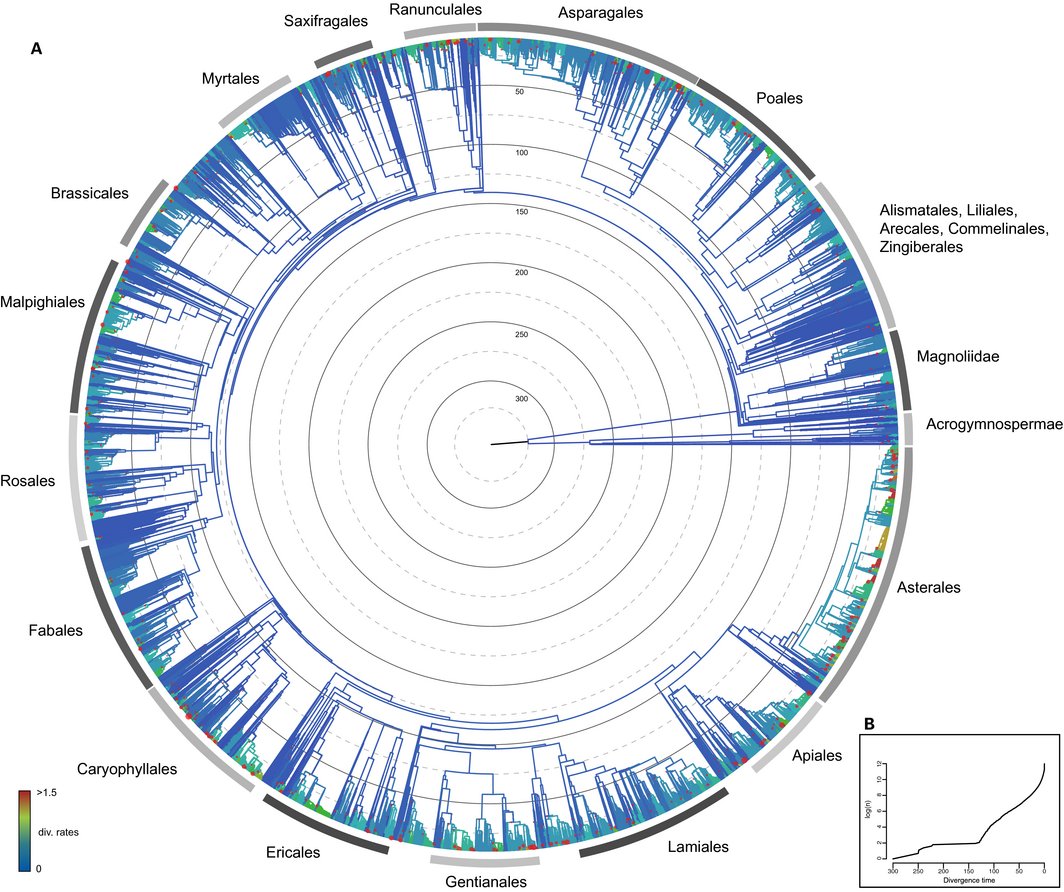
A couple of our papers just came out in the Tree of Life special edition of AJB. Pease et al. Quartet Sampling distinguishes lack of support from conflicting support in the green plant tree of life looks at different ways to examine support in a phylogeny using a bunch of green plant examples. Smith and Brown Constructing a broadly inclusive seed plant phylogeny uses OpenTreeOfLife and GenBank to make a big dated seed plant phylogeny.
-
Postdocs moving on to new positions
Congrats to Simon and Joseph who are moving on to a research position (Joseph) and a new phylogenetic startup phylagen.com (Simon). It was awesome to have you here at Michigan and good luck!


-
A number of new publications!
It has been a bit since our last update but just added a bunch of new papers to the publications page. This includes a first author publication by graduate student Joseph Walker! Congrats! Here is the list for the year so far
- Smith, S. A. and J. W. Brown. accepted. Constructing a comprehensive seed plant phylogeny.American Journal of Botany
- Pease J.B., J. W. Brown, J. F. Walker, C. E. Hinchliff, S. A. Smith. in press. Quartet Sampling distinguishes lack of support from conflicting support in the plant tree of life. American Journal of Botany
- Fleischmann, A., J. Schlauer, T. Givnish, and S. A. Smith. In press. Evolution of carnivory in angiosperms. Chapter 3. in forthcoming book on Carnivory in angiosperms.
- Lopez-Nieves, S., Y. Yang, T. Feng, S. A. Smith, S. F. Brockington, and H. A. Maeda. In press. Relaxation of Tyrosine Pathway Regulation Underlies the Evolution of Betalain Pigmentation in Caryophyllales. New Phytologist
- Brown, J. W., N. Wang, and S. A. Smith. 2017. The development of scientific consensus: analyzing conflict and concordance among Avian phylogenies. Molecular Phylogenetics and Evolution https://doi.org/10.1101/123034
- Brown, J. W. and S. A. Smith. 2017. The Past Sure Is Tense: On Interpreting Phylogenetic Divergence Time Estimates. Systematic Biology https://doi.org/10.1101/113720
- Walker, J., Y. Yang, M. Moore, S. F. Brockington, and S. A. Smith. 2017. Conflict among carnivores: whole transcriptomes unable to resolve the carnivorous clade of Caryophyllales. American Journal of Botany. https://doi.org/10.1101/115741
- Smith, S. A., M. J. Moore, S. F. Brockington, and Y. Yang. 2017. Disparity, Diversity, and Duplications in Caryophyllales. New Phytologist. https://doi.org/10.1101/132878
- Yang, Y., M. J. Moore, S. F. Brockington, J. Mikenas, and S. A. Smith. 2017. Improved transcriptome sampling pinpoints widespread paleopolyploidy events in Caryophyllales New Phytologist. https://doi.org/10.1101/143529
- Brown, J., J. Walker, and S. A. Smith. 2017. phyx: Phylogenetic tools for Unix. Bioinfor- matics. https://doi.org/10.1093/bioinformatics/btx063
- Yang, Y., M. J. Moore, S. F. Brockington, A. Timoneda-Monfort, T. Feng, H. E. Marx, Joseph F. Walker, and S. A. Smith. 2017. An efficient field and laboratory workflow for plant phylotranscriptomic projects. Applications in Plant Sciences. https://doi.org/10.1101/ 079582